yamanaka lab
cardiovascular disease
Understanding the Molecular Details of Pluripotency
Home
Areas of investigation
The goal of our laboratory is to understand the mechanisms underlying pluripotency and transcription factor-induced somatic cellular reprogramming. We have studied not only how cell fate can be changed but also the roles of RNA binding proteins such as LIN41 and NAT1/eIF4G2 in mouse and human pluripotent stem cells. Thorough the analyses of these RNA binding proteins, we concluded that post-transcriptional regulation is important for the maintenance of pluripotency and precise differentiation. Recently we have started to expand our research for the discovery of novel translation mechanisms and the full understanding of the mechanisms of translation initiation. Thus, the future direction of our research is trying to understand the specific and common roles of each eIF4G family in translation process.
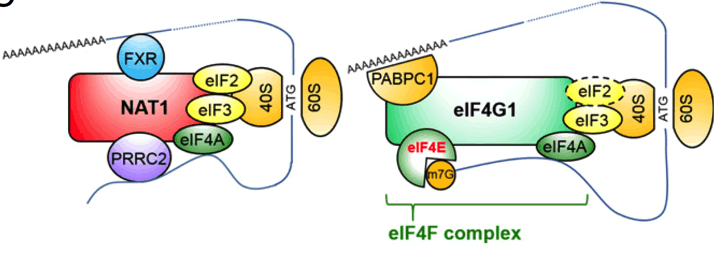 Models of cap-dependent translation with eIF4G1 and cap-independent translation with NAT1/eIF4G2. The dashed circle indicates a very small amount of eIF2 bound to eIF4G1 (Sugiyama et al, 2017).
Models of cap-dependent translation with eIF4G1 and cap-independent translation with NAT1/eIF4G2. The dashed circle indicates a very small amount of eIF2 bound to eIF4G1 (Sugiyama et al, 2017).
Significance
Textbooks have said that eukaryotic translation initiation factors gamma 1 (eIF4G1), one of eIF4G family, plays major roles in the initiation of protein translation in eukaryotic cells. However, our laboratory hypothesizes that other eIF4Gs such as NAT1 (also known as eIF4G2) and eIF4G3 have their important roles because at least NAT1 expression is much higher than other eIF4Gs in various tissues and cell types. Actually, the result of loss-of-function of NAT1 gene in mouse embryogenesis and ESCs suggested the essential roles of NAT1 in mouse early embryonic development and differentiation. In this period, we could identify the NAT1-interacting proteins. Some of them such as eIF3 and eIF4A were common in eIF4G1 and NAT1. However, our result also suggested that NAT1 has unique interacting partners such as FXR and PRRC2. These data encourages us to think about the unique role of NAT1 in non-canonical-translation rather than canonical cap-dependent translation driven by eIF4G1. Through the functional analyses of all eIF4Gs in parallel, we expect to find novel translation mechanisms such as cap-independent translation.
Approaches
Using iPSC lines derived from Fibrodysplasia ossificans progressiva (FOP) patients carry a missense mutation in ACVR1 (R206H) that leads to hyperactivation of BMP-SMAD signaling, we demonstrated that the BMP-SMAD-ID axis promoted human cellular reprogramming toward pluripotency by inhibiting p16INK4A expression.
We previously showed that LIN41 improved human cellular reprogramming, but the mechanisms had been unclear. We found novel roles of MYC and LIN41 in early stage of human cellular reprogramming: post-transcriptional activation of LIN41 expression by MYC is crucial for efficient expansion of reprogramming cells.
Twenty years ago, we found that NAT1-deficient mouse pluripotent stem cells impaired differentiation potentials, but the mechanism was unclear. We revisited the function of NAT1 in mouse pluripotency, and identified NAT1’s target gene which can explain the phenotype of NAT1-deficiency. Also, we identified the NAT1-interacting proteins using immunoprecipitation and mass spectrometric analyses.
 Yamanaka Lab Group 2021
Yamanaka Lab Group 2021



